- About the Structural Kinome
The Structural Kinome Site is a centralized platform dedicated to the structural and functional exploration of the human kinome. Our mission is to integrate, visualize, and analyze the kinase three-dimensional structures to advance understanding of their catalytic mechanisms, conformational diversity, and ligand interactions. By curating experimentally determined and computationally modeled kinase structures, it serves as a bridge between structural biology, databases, AI models, and drug discovery. Researchers can use it to identify active and inactive inhibitors, explore covalent and non-covalent binding sites, and compare structural features across kinase families, supporting applications in kinase inhibitor design, polypharmacology profiling, and disease-associated target analysis, offering a foundation for systematic, structure-guided studies in chemical biology and precision medicine.
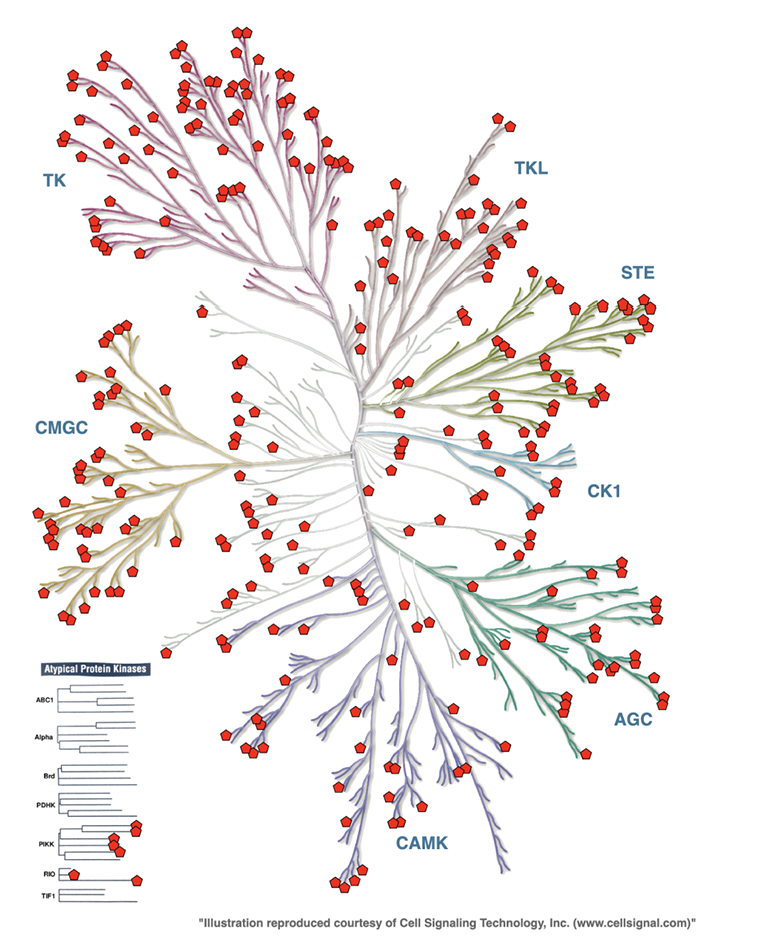
Statistics (Updated on June 28, 2025)
| Statistics | Number |
|---|---|
| Structures(PDB IDs) | 6969 |
| Covered Kinases | 355 (Human: 311; Mouse: 42; Chicken: 2) |
| Kinase-ligand Complexes (PDB IDs) | 6122 |
| Apo Kinase structures (PDB IDs) | 847 |
| Covalent-Inhibitor-binding structures (PDB IDs) | 321 |
Recent Updates
1. Kinase Structures
2. Small molecules inhibitors
3. Degrader
4. ...
Reference
We will add the papers once published
School of Data Science, University of Virginia, Charlottesville, Virginia 22904
Email: zz7r@virginia.edu